Project 3: Weakly and semi-supervised learning for segmentation of volumetric medical images with fewer training examples
Background
Advanced machine learning technologies have revolutionized image analysis in the recent years. In various vision tasks, current methods that utilize multi-layered neural networks, a.k.a. deep learning, reach near-human performance. Tasks related to medical image computing have also seen a surge in performance with the introduction of deep learning techniques. In particular, performance of DL-based algorithms in automatic segmentation and detection of normal structures and lesions have reached a level where these methods can accelerate clinical workflow in radiology and radiation oncology, and enable high-throughput imaging-based clinical research. One of the most important components that lead to DL’s recent success is the availability of large-scale labeled datasets. DL algorithms have large number of parameters that allow them to represent complex patterns of statistical dependence between explanatory and outcome variables. In segmentation of medical images, the labeled examples needed for training DL algorithms consists of images showing the structure of interest and corresponding pixel-wise annotations delineating the boundaries of the structure. The resource requirements for creating large labeled datasets create an obstacle for applying DL algorithms on new problems.
Working Hypothesis
- DL-based segmentation methods can be trained using “weak” labels in the form of scribbles rather than detailed boundary delineations and this strategy would require less resource commitments in terms of number of expert-hours for creating datasets of training examples.
- Very accurate DL-based medical image segmentation methods can be trained using few labeled and large number of “un-labeled” examples, where only the image is available.
Specific aims for this research project
- Develop novel algorithms to train DL-based segmentation methods for volumetric medical images using weak segmentation labels.
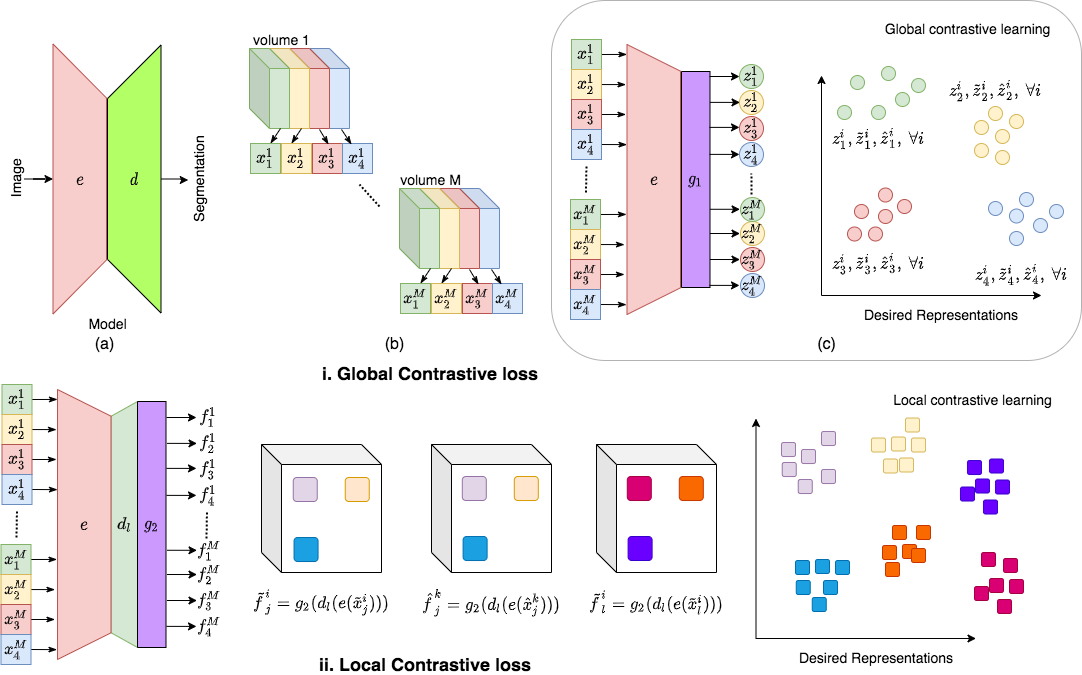
- Develop novel semi-supervised / self-supervised volumetric segmentation methods that utilizes unlabeled images along with few labeled examples. This is illustrated in below figure.
- Investigate application of the developed techniques for segmenting brain tumors in MRI (relation to WP2 and WP7), lymph nodes and stations in the head-and-neck area (relation to WP5).
Highlighted recent work
We are happy to announce that parts of our work was accepted to NeurIPS as an oral presentation!
(as one of only 109 out of 9000 submissions)
Title:
Contrastive learning of global and local features for medical image segmentation with limited annotations
Abstract:
A key requirement for the success of supervised deep learning is a large labeled dataset - a condition that is difficult to meet in medical image analysis. Self-supervised learning (SSL) can help in this regard by providing a strategy to pre-train a neural network with unlabeled data, followed by fine-tuning for a downstream task with limited annotations. Contrastive learning, a particular variant of SSL, is a powerful technique for learning image-level representations. In this work, we propose strategies for extending the contrastive learning framework for segmentation of volumetric medical images in the semi-supervised setting with limited annotations, by leveraging domain-specific and problem-specific cues. Specifically, we propose (1) novel contrasting strategies that leverage structural similarity across volumetric medical images (domain-specific cue) and (2) a local version of the contrastive loss to learn distinctive representations of local regions that are useful for per-pixel segmentation (problem-specific cue). We carry out an extensive evaluation on three Magnetic Resonance Imaging (MRI) datasets. In the limited annotation setting, the proposed method yields substantial improvements compared to other self-supervision and semi-supervised learning techniques. When combined with a simple data augmentation technique, the proposed method reaches within 8% of benchmark performance using only two labeled MRI volumes for training.
Here is the arxiv link to the article.